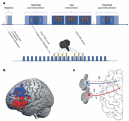
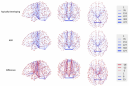
ABOUT PENNSIVE
The Penn Statistics in Imaging and Visualization Endeavor (PennSIVE) consists of a group of statisticians studying etiology and clinical practice through medical imaging.
LOCATION
2nd Floor, Blockley Hall
University of Pennsylvania
423 Guardian Drive
Philadelphia, PA 19104-6021